
Genomics Specialist
Contact Krista
Public health officials and decision makers in the biothreat community must anticipate emerging biological threats by proactively evaluating DNA and RNA sequences. For public health officials deployed to areas of suspected outbreaks, portable nanopore sequencing hardware enables the rapid identification and characterization of biological threats in field operations. Nanopore sequencing also offers a lower initial investment cost, has the potential to report sequence data analysis results in real-time, and can capture more of a microbial genome within a single, long read. Further, advances in bioinformatics now offer the biosurveillance community a capability to recognize new, emerging, or minimally characterized biological threats from gene sequence data.

Analysis of Simulated Novel Pathogens

SeqScreen-Nano Development
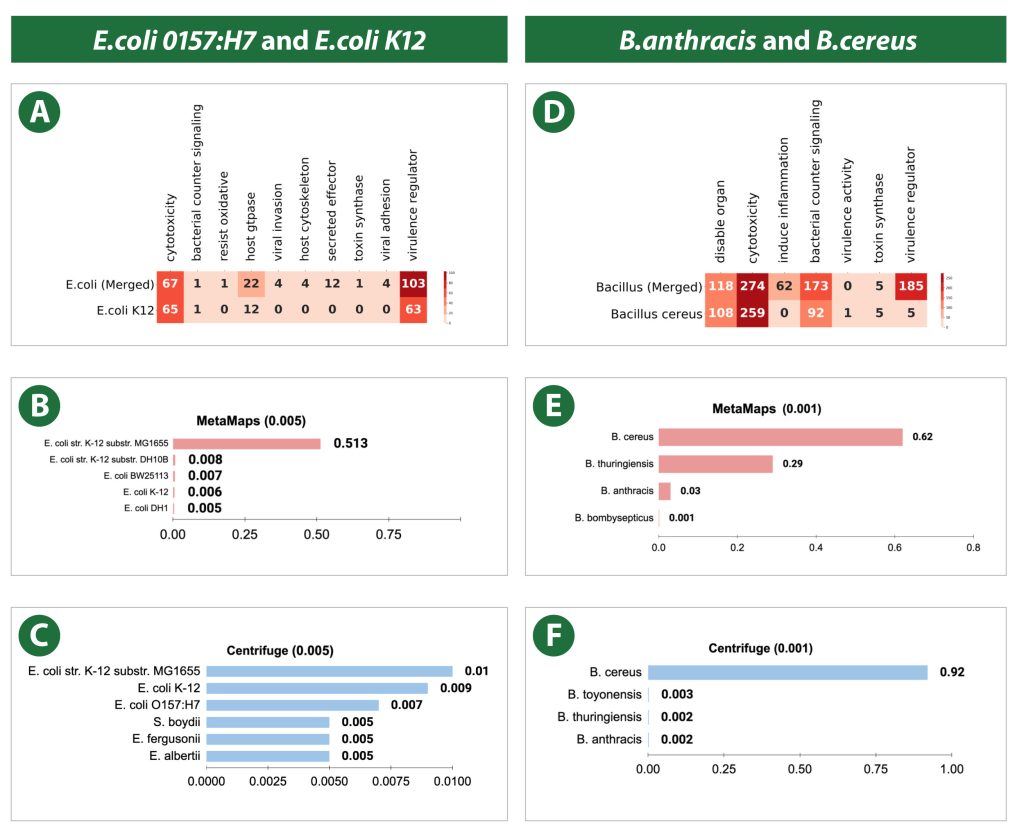
SeqScreen-Nano was developed by Signature Science and Rice University with funding from IARPA’s Fun GCAT program. This tool enables biothreat assessors’ analysis of nanopore sequencing results in real-time from portable sequencing hardware. SeqScreen-Nano goes beyond taxonomic (genus-species)
identification of known pathogens to also report functional threat assessments for both known and emerging pathogens. The software performs analyses in real time, directly from Oxford Nanopore Technologies’ MinION sequences in simple or complex samples (Balaji et al. 2023). Using this bioinformatics software tool, biothreat assessors can now determine the threat potential of individual gene sequences based on their pathogenic function, a capability that revolutionizes biothreat detection by delivering real-time assessments of unknown or emerging threats (Balaji et al. 2022).
For field forward threat assessors using nanopore sequencing, data analysis for real-time threat assessment can amplify the value of in-field sequencing by enabling timely decision making in critical situations. SeqScreen-Nano marks a significant step toward that goal.
Publications
Want more information about SeqScreen-Nano?
